Radical roadmap: Russell lab develops novel technique to analyze the host-pathogen dynamic
Tuberculosis continues to be a deadly threat across the globe, and our knowledge regarding what controls disease progression is limited. Now, in a paper published July 22 in the Journal of Experimental Medicine, a group at the Cornell College of Veterinary Medicine has developed a novel method that analyzes how individual immune cells react divergently to the bacteria that cause tuberculosis. Not only could this pave the way for new vaccine strategies against this deadly disease, but the technology could be used with other infectious pathogens, providing new insights into infectious diseases around the world.
For years, the lab of Dr. David Russell, the William Kaplan Professor of Infection Biology in the Department of Microbiology and Immunology, has developed a series of cutting-edge technologies to unravel how Mycobacterium tuberculosis (Mtb), the bacteria that cause tuberculosis, infect and persist in their host cells — which are typically immune cells called macrophages.
Their latest innovation combines two analytical tools that each target a different side of the pathogen-host relationship: “reporter” Mtb bacteria that glow different colors depending on how stressed they are in their environment, and single-cell RNA sequencing (scRNA-seq), which yields RNA transcripts of individual host macrophage cells.
“For the first time ever, Dr. Davide Pisu in my lab combined these two approaches to analyze Mtb-infected immune cells from an in vivo infection,” Russell said.
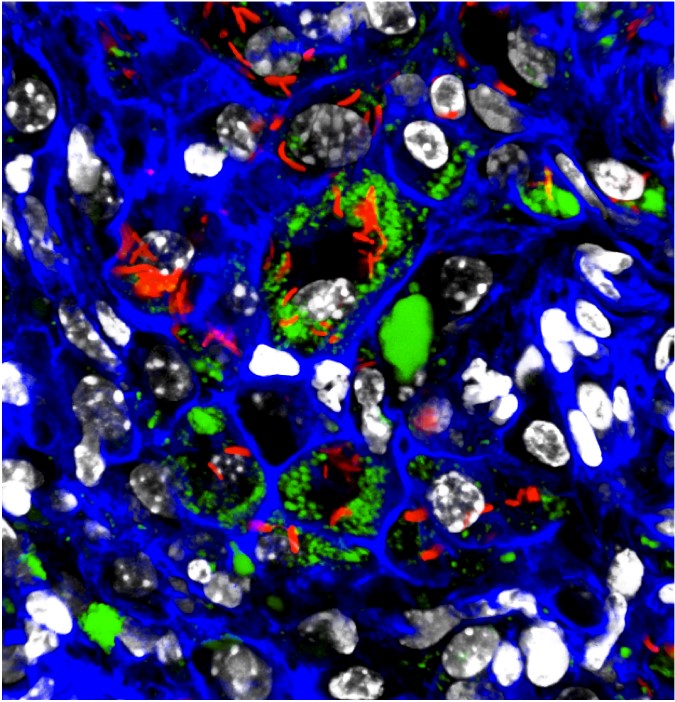
After infecting mice with the fluorescent reporter Mtb bacteria, Russell’s team was able to gather and flow-sort individual Mtb-infected macrophages from the mouse lung, and determined which macrophages promoted Mtb growth (sporting happy, red-glowing bacteria) or contained stressed Mtb unlikely to grow (unhappy, green-glowing bacteria).
Next, they took the two sorted, infected macrophage populations and ran them through single-cell RNA sequencing analysis, thereby generating transcriptional profiles of each individual host cell in both populations.
A nearly perfect match
When the scientists compared the macrophage single-cell sequencing data with the reporter bacteria phenotype, they found an almost perfect one-to-one correlation between the fitness status of the bacterium and the transcriptional profile in the host cell. Macrophages that housed unhappy green bacteria also expressed genes that were known to discourage bacterial growth, while those with happy red bacteria expressed genes known to promote bacterial growth.
While one might expect such data to match up naturally, scientific experiments rarely play out so nicely. “What absolutely stunned us is how well it worked,” said Russell. “When Davide Pisu showed me the analysis I nearly fell off my chair.”
Normally, phenotypes and transcriptional profiles are two characteristics that rarely come together in a perfect match, and this almost never happens from in vivo data.
This near perfect match up of bacterial fitness and transcriptional profiles revealed new nuances. “While our previous results identifying the resident alveolar macrophages (AM) as permissive and the blood monocyte-derived recruited macrophages (IM) as controlling Mtb infection was correct in a broad sense, we found, unsurprisingly, that this was an oversimplification,” said Russell.
There was variation even within these two different macrophage types: Some AM cells controlled Mtb growth while some IMs were allowing bacterial expansion. The team found that comparable subsets of immune cells were present in both human and mouse lung samples.
An additional step in the study was to look at whether the responses of AM and IM cells to Mtb were epigenetically controlled. This could explain how two sets of macrophages respond divergently to the same immune milieu. Using ATAC-seq, which provides a read-out of a cell’s epigenetic landscape, they found that this was the case.
“The analysis showed that when these cells are exposed to Mtb or the vaccine strain (BCG) — through infection or vaccination, respectively — their epigenetic programming has a major influence over whether their response leads to disease control or progression,” said Russell.
New roadmap
Armed with this body of new information, the Russell lab plans to hit the ground running to identify novel therapeutics. “We’re going to begin by screening libraries of known epigenetic inhibitor compounds to see which ones might be useful in modifying the immune response,” said Russell.
If they do find promising compounds — ones that push macrophages towards an anti-Mtb behavior — they could potentially be used in combination with vaccines to assist a patient’s immune system in protecting against tuberculosis.
The finding now lays a foundation for much more powerful studies on how pathogens affect individual cells, allowing for a holistic examination of the system. “This is a roadmap that lets us look across an entire population of cells and see how a single perturbation impacts the cells across that population,” said Russell. “We can test for drug efficacy in in vivo infection without any preconceived limitation on how compounds may function.”
This approach is extremely flexible and could be used in the study of any intracellular pathogen, including viruses, and is readily applicable to any animal challenge model.
Written by Lauren Cahoon Roberts